DNA Techniques Applied to the Identification of Pinctada Fucata Pearls from Uwajima, Ehime Prefecture, Japan

ABSTRACT
The genetic material deoxyribonucleic acid (DNA) is a useful indicator in identifying pearl species, provided the extraction and amplification of a targeted component is possible from amounts of pearl powder small enough to render the technique essentially nondestructive. This study builds on similar work carried out by Meyer et al. (2013) but focuses on akoya cultured pearls from Uwajima in Ehime Prefecture, Japan. Here, we also target the 16S rRNA genes, the mitochondrial component of DNA to code 16S rRNA (ribosomal ribonucleic acid). We successfully amplified the 16S rRNA gene from the mantle tissues of donor and host pearl oysters and from 5–10 mg of pearl powder samples. The results indicate a match with the Pinctada fucata 16S rRNA gene.
INTRODUCTION
Modern pearl identification—including the type of pearl and any treatment applied—is performed by applying almost exclusively nondestructive methods. These include macro to microscopic observations, spectroscopy, chemical analysis, fluorescence, and various forms of X-ray imaging (Scarratt, 2011). Surface inspection differentiates nacreous from non-nacreous pearls, while chemical analyses using X-ray fluorescence and luminescence can distinguish between saltwater and freshwater pearls (e.g., Hänni, 2000; Hänni et al., 2005; Kessrapong et al., 2017). Internal inspection using X-radiography, which was introduced to gemology in the 1930s (Dirlam and Weldon, 2013), has evolved into high-tech X-ray computed microtomography (μ-CT). This technology enables more consistent separation of natural, non-bead-cultured, and bead-cultured pearls (e.g., Sturman, 2009; Karampelas et al., 2010).
One of the ongoing challenges is to identify the saltwater mollusk species producing white, cream, or silver nacreous pearls, since these can form in all Pinctada species as well as some other species, and can have similar appearances. As pearls are formed by mollusks, a direct identification method would be to find biological evidence such as DNA indicators. (For boldfaced terms, please refer to the glossary.
DNA studies emerged in the biological sciences (e.g., Toro, 1998; Masaoka and Kobayashi, 2004; Freier et al., 2008) after the development during the 1980s of the polymerase chain reaction (PCR) method of duplicating gene fragments (see figure 1A). A number of studies on pearl oysters of the Pinctada species (e.g., Masaoka and Kobayashi, 2006; Tëmkin, 2010; Masaoka et al., 2016) have investigated their nuclear and mitochondrial ribosomal ribonucleic acid (rRNA) and the internal transcribed spacer (ITS) regions of nuclear rRNA genes while discussing the phylogeny and identification of the species.
According to these studies, pearl oysters can be classified into three groups—(1) P. maxima, (2) P. margaritifera, and (3) akoya-type—that have different nuclear 28S rRNA and mitochondrial 16S rRNA genes. Akoya-type oysters include P. fucata (Indo-Pacific region), P. radiata (eastern Indian Ocean to Red Sea), and P. imbricata (western Atlantic region), that have the same nuclear 28S rRNA gene but different mitochondrial 16S rRNA genes (Masaoka et al., 2016). Thus, DNA investigation has demonstrated that the members of the akoya pearl oyster group are genetically very closely related species but distinct genealogical units, suggesting subspecies-level differences (Tëmkin, 2010). Tëmkin also pointed out that it might be meaningful to treat the three individual akoya populations as evolutionary significant units, because they originated from three different geographic regions. 16S rRNA genes differ in all five of the most abundant pearl oysters (P. maxima, P. margaritifera, P. fucata, P. imbricata, and P. radiata), making them very useful DNA markers to identify the Pinctada species or subspecies.
The targeted component of the DNA, the 16S rRNA gene, is the component necessary to code the 16S rRNA, essential in the process of biological protein synthesis. Because the cells of higher-order animals are known to have many copies, the DNA regions have the advantage of developing sensitive PCR-based identification methods (Masaoka and Kobayashi, 2005). Furthermore, 16S rRNA genes are conserved within a Pinctada species but contain sufficient variation between species to produce species-specific sequences (Masaoka and Kobayashi, 2004, 2005). These sequences have been registered in GenBank, the genetic sequence database of the National Institutes of Health’s National Center for Biotechnology Information (NCBI).
Using a micro-drilling method, Meyer et al. (2013) succeeded in extracting nuclear and mitochondrial DNA samples and identifying the pearl oyster species as P. margaritifera, P. maxima, or P. radiata. For the present study, akoya cultured pearl oysters were sampled during harvesting (hamaage) in Uwajima, in Japan’s Ehime Prefecture, with the purpose of extracting DNA and the mitochondrial 16S rRNA gene from minute amounts of pearl powder and tissues of P. fucata. (For the history of pearl culturing in Uwajima, see box A.) Our goal was to conclusively identify the species and use that as a reference for determining the whole targeted DNA sequence. The Japanese akoya pearl oysters P. martensii and P. fucata martensii have been distinguished from P. fucata based on the classical biological classification such as shell morphologies in most cases (e.g., Hayami, 2000; Masaoka and Kobayashi, 2004). However, their genetic properties are very similar, with the same 16S rRNA genes, indicating that P. fucata and P. martensii are genetically much closer to other akoya-type species such as P. radiata and P. imbricata (Masaoka and Kobayashi, 2004; Wada and Tëmkin, 2008). This study treats P. martensii as a subspecies of P. fucata, as their 16S rRNA genes have the same DNA sequence alignments.
MATERIALS AND METHODS

The shells sampled for this project were collected from live pearl oysters in January 2016 at Uwajima (see the harvesting details in figure 2). Thirty-seven pearl oysters, consisting of 28 host oysters used to grow pearls and nine donor oysters bred for tissue grafting, were collected after they were confirmed to be alive and immediately transported to the University of Tokyo. The shells were dissected, and both the mantle and pearl samples were frozen and stored at –80°C until analysis was possible. For DNA extraction experiments, the mantle tissue of a host oyster and a donor oyster were selected together with four pearls (see table 1).
 Supplementary figure S-1
Supplementary figure S-1
Our strategy was to use a simple direct sequence method consisting of three experimental steps: (1) DNA extraction and subsequent purification, (2) amplification and detection, and (3) sequencing. For the first steps of DNA extraction and purification, 16S rRNA gene extraction from mantle tissues was performed using a general phenol-chloroform extraction process (see Suzuki et al., 2004), whereas 16S rRNA genes were obtained from pearl powder samples using a method similar to that described in Meyer et al. (2013). The second step of DNA amplification consists of the PCR and nested PCR methods (figure 1A). The PCR method consists of three repeating processes: denaturation, annealing, and extension using the PCR machine. Denaturation is separating the targeted DNA double strands, called “DNA templates,” into two single strands. Annealing is a connecting process of the single DNA strands with forward or reverse primers, which are artificially aligned short nucleotides (about 10–15 nucleotides). Extension is extending the double chain of the single DNA strands with available nucleotides, called deoxynucleotides (see figure 1B). These three processes are repeated for 30–40 cycles, and targeted DNA (the 16S rRNA gene) should be amplified exponentially. The nested PCR method is known to improve PCR sensitivity for specific DNA amplification and is often used for complementary DNA cloning (e.g., Suzuki et al., 2004; Liu et al., 2007), as well as to recover ancient DNA (e.g., Salo et al., 1994; Yang et al., 1997). Meyer et al. (2013) also applied the nested PCR method after drilling pearl samples to obtain as little as 10 mg of powder and succeeded in amplifying nuclear ITS2 regions (refer to glossary for the definition of ITS). The determination of whether DNA was amplified was performed using a gel electrophoresis method (figure 1B). Negatively charged DNA molecules migrate though the gel when an electric field is applied and are separated based on their size. Thus this method produces the quantity and approximate molecule size of amplified DNA necessary for the third step of sequencing. We applied the Sanger method to determine the nucleotide arrangement (figure 1C), added fluorescent-labeled dideoxynucleotides to amplify the DNA template, and subsequently generated DNA fragments as the dideoxynucleotides terminated the DNA extension process (figure 1C, left). The DNA fragments were separated by capillary electrophoresis on the basis of the size, producing the electropherogram as the final sequence result (figure 1C, right). The detailed DNA extraction to detection methods are described in box B.
RESULTS AND DISCUSSION
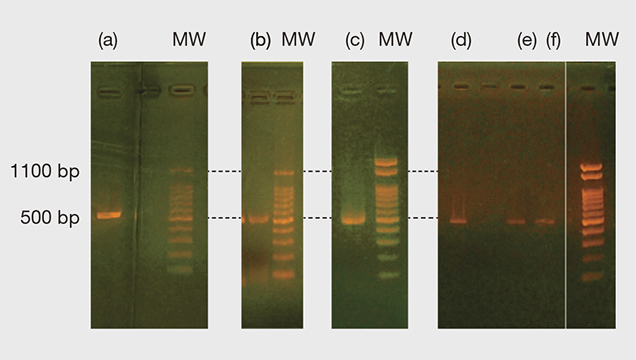
Figure 3 shows the PCR and nested PCR products of the 16S rRNA gene obtained from the gel electrophoresis process. All the bands appeared at approximately 500 base pairs (bp) of molecular weight markers, and the resulting forward and reverse sequences mostly matched with 524 bp of P. fucata (see table 1 and supplementary figure S-1. For one pearl sample (no. 20) we only obtained the forward sequence with 820 bp in total. However, the part of the sequence with 350 bp is perfectly matched within the part of the 524 bp range expected for P. fucata and not with sequences of the other Pinctada species on record, including P. radiata and P. imbricata (see supplementary figure S-2). Thus, all the 16S rRNA gene samples obtained matched P. fucata.
In this study, it was possible to extract DNA from 5 to 10 mg of pearl powder, but the pearls were severely damaged in the process (see figure B-1, right column). To apply the DNA methods effectively within the pearl industry, further improvements to the extraction step are clearly needed. Reducing the sample size required to approximately 0.01 mg would be one such improvement. This would equate to the approximate amount ablated during laser ablation–inductively coupled plasma–mass spectroscopy (LA-ICP-MS) chemical measurements (Hänni and Cartier, 2013). DNA amplification using PCR was improved by (1) the application of the nested PCR method and (2) the use of two different types of PCR polymerase: Taq-type and KOD-type (summarized in table 1).
A powder sample was obtained from pearl sample no. 14 to successfully amplify the 16S rRNA using the nested PCR method with a Taq-type polymerase similar to that applied by Meyer et al. (2013). The DNA of the other three pearl samples was amplified using only the first (non-nested) PCR with a KOD-type polymerase. The advantage of KOD-type polymerase is that it has 3'→5' exonuclease activity (Morikawa et al., 1994), which is an active process mechanism of “proofreading” the polymerase chain reaction. The Taq polymerase does not have this proofreading activity. At present it is not known why different polymerase activities for the pearl samples occurred, but they may help amplify 16S rRNA genes in various pearl powder samples.
Future research needs to address the effects of any treatments, such as maeshori or bleaching, on DNA extraction. Although the species and distinction of the akoya-type pearl oysters are still under discussion, the draft genome of P. fucata has been provided by Takeuchi et al. (2012) and ongoing DNA studies will continue. Future results will aid in species identification and may also provide evidence of the geographic region where pearls formed, benefiting the pearl industry in matters of traceability.
CONCLUSIONS
The 16S rRNA genes extracted from the mantle tissues of one donor shell and one host shell fully matched the complete sequence of P. fucata. The 16S rRNA genes extracted from the powder of the nacreous layers of four bead-cultured pearl samples were also found to match the sequence of P. fucata. Thus, the pearl oysters and pearl samples obtained in January 2016 from Uwajima were categorically identified as originating from P. fucata species. From a methodology perspective, it was possible to identify the P. fucata 16S rRNA gene from only 5 to 10 mg of pearl powder sample weight, compared to the 10 to 100 mg quantity previously reported by Meyer et al. (2013). This reduced sampling is important when analyzing pearls of historical significance and value.
GLOSSARY
| Base pair (bp): A pairing of the molecules that comprise DNA, specifically an adenine (A)/thymine (T) or guanine (G)/cytosine (C) pair. Deoxyribonucleic acid (DNA): The hereditary material in humans and almost all other organisms. Most DNA is located in the cell nucleus, but a small amount can also be found in the mitochondria. Complementary DNA is DNA synthesized by reverse transcription reaction of messenger RNA. Dideoxynucleotide: A type of nucleotide that terminates the DNA extension process, as it does not contain the OH– (hydroxyl) groups needed for extending DNA chains. Electropherogram: Diagram produced by electrophoresis. Exonuclease activity: An activity of an enzyme to hydrolyze a DNA chain sequentially from the end of a polynucleotide molecule. Evolutionary significant unit: A population of subspecies organisms that is considered distinct for conservation purposes. Proposed by Ryder (1986). Gene: The basic physical and functional unit of heredity. A “gene” is encoded by DNA and usually acts as a template to make a protein. Internal transcribed spacer (ITS): The gene regions between nuclear rRNA genes. Koshi-mono: The Japanese term for akoya cultured pearls removed from mollusks 12 months or more after seeding. Mitochondrion: An organelle in cells that have a nucleus, as in plants and animals. Organelle: Small, specialized structures in cells that operate like organs by carrying out specific tasks. Polymeric: Referring to an aggregate of monomers or combined molecules. Protein: A molecule consisting of amino acids that carries out functions related to an organism’s biology. Phylogeny: The history of the evolution of a species. Ribonucleic acid (RNA): A polymeric molecule involved in various biological roles for coding, decoding, regulation, and expression of genes. 16S rRNA (16S ribosomal RNA) is a component of ribosomal RNA, and its gene is coded in mitochondria. Tonen-mono: The Japanese term for akoya cultured pearls removed from the mollusks within a year of seeding. |





